Small-plot analysis
small_plot_analysis.Rmd
library(CBADASReml)
#> Loading required package: asremlPlus
#> ASReml-R needs to be loaded if the mixed-model functions are to be used.
#>
#> ASReml-R is available from VSNi. Please visit http://www.vsni.co.uk/ for more information.
#> Loading required package: ggplot2
#> Loading required package: gstat
#> Loading required package: sf
#> Linking to GEOS 3.13.0, GDAL 3.8.5, PROJ 9.5.1; sf_use_s2() is TRUE
library(asreml)
#> Loading required package: Matrix
#> Online License checked out Fri Aug 22 12:51:01 2025
#> Loading ASReml-R version 4.2CBADASReml
Introduction
A typical type of experimental analysis that this package will assist with is the small plot trial. Here we will show the general workflow for performing a small-plot analysis and how each function from the package can be used. Analysis of small plot trials is done using linear mixed-effects models (LMMs), using {ASReml} or {glmmTMB}, and this package contains helper functions for this type of modelling.
Note that the trial design is not handled at all by this package, and we recommend the use of {speed} or {FielDHub} for creating small plot trial designs.
Mathematical Background
Let:
Where
- is the vector of observations.
- is the vector of fixed effects.
- is the design matrix for the fixed effects.
- is the vector of random effects.
- is the design matrix for the random effects.
- is the vector of residuals.
It is assumed that the vectors and are independent, and their joint distribution is a multivariate Gaussian described by:
Where and are covariance matrices for and and are functions of the variance parameters and , and finally the dependent variable is described by:
These and covariance matrices, or covariance structures, are very important. These represent the way the random effects and the residuals behave, and and , the covariance structure parameters are the things we try and estimate when we use ASReml.
In the case of independent and identically distributed (IID) residual structures, the residual covariance parameter will be some variance component .
Exploratory Data Analysis
After gathering data from an experiment, it is useful to perform an
exploratory data analysis on the raw data, and for this you will likely
use a combination of tidyverse and gplot2 for
this, to visualise outliers, the data distribution, etc. in order to get
a preliminary grasp of what you are working with, and to observe the
data for any issues that will hinder the actual analysis. As an
assistance with this process, the function
exploratory_table is provided.
CBADASReml::exploratory_table(
data = oats,
response = "yield",
groupby = c("Variety", "Nitrogen")
)
#> Variety Nitrogen mean median sd
#> 1 Golden_rain 0_cwt 80 75 21.0047613649858
#> 2 Golden_rain 0.2_cwt 98.5 102.5 13.4721935853075
#> 3 Golden_rain 0.4_cwt 114.666666666667 110 29.9443929086343
#> 4 Golden_rain 0.6_cwt 124.833333333333 129.5 20.8750249500849
#> 5 Marvellous 0_cwt 86.6666666666667 92.5 16.5730705262081
#> 6 Marvellous 0.2_cwt 108.5 111.5 26.8533051969399
#> 7 Marvellous 0.4_cwt 117.166666666667 118.5 9.78604448521805
#> 8 Marvellous 0.6_cwt 126.833333333333 122.5 20.2920345620311
#> 9 Victory 0_cwt 71.5 65 20.59854363784
#> 10 Victory 0.2_cwt 89.6666666666667 89.5 22.5092573548455
#> 11 Victory 0.4_cwt 110.833333333333 106 26.0108951531213
#> 12 Victory 0.6_cwt 118.5 114.5 30.0915270466622
#> cv reps min max range
#> 1 26.2559517062322 6 60 117 60 - 117
#> 2 13.6773538937132 6 82 114 82 - 114
#> 3 26.1142961412508 6 86 161 86 - 161
#> 4 16.7223163819105 6 96 149 96 - 149
#> 5 19.1227736840862 6 63 105 63 - 105
#> 6 24.7495900432626 6 70 140 70 - 140
#> 7 8.35224280388454 6 104 132 104 - 132
#> 8 15.9989760016014 6 99 156 99 - 156
#> 9 28.809151941035 6 53 111 53 - 111
#> 10 25.103260990534 6 64 130 64 - 130
#> 11 23.4684768298838 6 81 157 81 - 157
#> 12 25.3936937102634 6 86 174 86 - 174Model Fitting and Validation
We may use the R package asreml as follows to implement
an LMM. For the purpose of this vignette we will fit a few different
models, so we can demonstrate different techniques from the package.
# The needs to be ordered so that the model can run
oats <- oats[order(oats$Column), ]
mod_oats <- asreml(
fixed = yield ~ Variety + Nitrogen + Variety:Nitrogen,
random = ~ idv(Blocks) + idv(Blocks):id(Wplots),
residual = ~ ar1(Column):ar1(Row),
data = oats,
trace = FALSE
)
#> Warning in asreml(fixed = yield ~ Variety + Nitrogen + Variety:Nitrogen, : Some
#> components changed by more than 1% on the last iteration
mod_oats <- update(mod_oats, aom = TRUE)
mod_rats <- asreml(
fixed = weight ~ littersize + Sex + Dose,
random = ~idv(Dam),
residual = ~units,
data = rats,
trace = FALSE
)
mod_rats <- update(mod_rats, aom = TRUE)In the first model, mod_oats, we are using the
oats data:
Data
- The
oatsdata has 72 rows, 2 treatments (Nitrogen and Variety), and plots (3 levels) within blocks (6 levels). Therefore:- (number of observations)
- (number of fixed effects, including an interaction effect)
- (number of random effects, blocks, and plots within blocks)
-
BlocksandBlocks:Wplotsmake up our matrix. Since these are factors, each level is treated as it’s own dummy variable. We end up with two matrices which make up which are given as follows (if ordered by their respective variables):
Where each column in
represents the
Blocks, and the columns in
represent the
Blocks
Wplots.
- Since we have
model terms (
idv(Blocks)andidv(Blocks):id(Wplots)), we have two matrices, and also two vectors. Our random effects vector is given by:- .
-
idv(Blocks) + idv(Blocks):id(Wplots)is the random effects variance structure. Since we have two terms, we again have two ’s. We specify theidvvariance model function, which is the homogeneous scaled identity variance structure, i.e. . The second term has a /direct product/ implied by the:operator, which in the case of matrices is a /Kronecker product/, ofidv(Blocks)andid(Wplots).idvariance model means it is simply a correlation structure, i.e. , and there is no estimation of a variance parameter. Noting we have 2 random effects terms:- ( is the identity matrix)
- The error vector is specified by the single model term
ar1(Column):ar1(Row), therefore the error is given by:- where is the structure
-
ar1(Column):ar1(Row)specifies the residual variance structure. In this case we refer to an autoregressive model of order 1 (AR(1)) on bothRowandColumn, where the:operator implies a separable Kronecker product structure. The variance modelar1is of the form: .- Where refers to the correlation within the columns () or the rows ().
In the second model, mod_rats, we are using the
rats data:
-
weightis our vector. -
littersize,SexandDoseare in the matrix. -
Damis in the matrix. We specify theidvvariance model function, which is the homogeneous scaled identity variance structure, i.e. . In other words, the term is IID:-
is the design matrix for
Dam. - .
-
is the design matrix for
-
idv(units)is the residual variance structure, in this case the residual term has the homogeneous covariance matrix: . This is basically saying all observations have the same error distribution (IID):
Upon executing the above in R, the ASReml-R code will
attempt to estimate the variance components
and
,
using the REML algorithm.
Function examples
CBADASReml Diagnostics
outlier_summary
The purpose of the outlier_summary function is to
observe potential outliers that can then be discussed with the data
provider. Below is an example for the rats dataset that is included
within ASReml-R.
outlier_summary(mod_rats)
#>
#> ── Outliers detected: 2 ────────────────────────────────────────────────────────
#> Dose Sex littersize Dam weight units combined_trt residuals
#> 56 C F 13 5 5.02 56 13_F_C -4.090506
#> 66 C F 9 6 3.68 66 9_F_C -7.941340
#>
#> ── Data table ──
#>
#> Dose Sex littersize Dam weight units combined_trt residuals
#> 56 C F 13 5 5.02 56 13_F_C -4.0905065
#> 57 C F 13 5 6.04 57 13_F_C -1.4769901
#> 53 C F 13 5 7.16 53 13_F_C 1.3927533
#> 55 C F 13 5 7.14 55 13_F_C 1.3415079
#> 54 C F 13 5 7.09 54 13_F_C 1.2133944
#> 129 C F 13 10 5.92 129 13_F_C -1.1145276
#> 126 C F 13 10 6.67 126 13_F_C 0.8050510
#> 128 C F 13 10 6.53 128 13_F_C 0.4467297
#> 130 C F 13 10 6.52 130 13_F_C 0.4211353
#> 125 C F 13 10 6.44 125 13_F_C 0.2163802
#> 131 C F 13 10 6.44 131 13_F_C 0.2163802
#> 127 C F 13 10 6.43 127 13_F_C 0.1907859
#> 66 C F 9 6 3.68 66 9_F_C -7.9413397
#> 64 C F 9 6 7.26 64 9_F_C 1.3727706
#> 65 C F 9 6 6.58 65 9_F_C -0.3963900
directional_variograms
The purpose of this function is to view the autocorrelation structure
in both directions, rather than the traditional approach with
ASReml-R.
plot(directional_variograms(mod_oats))
Model Outputs
Traditional Outputs (ASReml-R)
wald
For getting the significance of each variable in the model
wald(mod_rats)
#> Wald tests for fixed effects.
#> Response: weight
#> Terms added sequentially; adjusted for those above.
#>
#> Df Sum of Sq Wald statistic Pr(Chisq)
#> (Intercept) 1 1477.67 8981.5 < 2.2e-16 ***
#> littersize 1 4.58 27.8 1.312e-07 ***
#> Sex 1 9.80 59.5 1.199e-14 ***
#> Dose 2 3.76 22.8 1.100e-05 ***
#> residual (MS) 0.16
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
predict
predict.asreml(mod_oats, classify = "Nitrogen")
#> $pvals
#>
#> Notes:
#> - The predictions are obtained by averaging across the hypertable
#> calculated from model terms constructed solely from factors in
#> the averaging and classify sets.
#> - Use 'average' to move ignored factors into the averaging set.
#> - The simple averaging set: Variety
#> - The ignored set: Blocks,Wplots
#>
#> Nitrogen predicted.value std.error status
#> 1 0_cwt 77.76388 6.929776 Estimable
#> 2 0.2_cwt 100.15097 6.896569 Estimable
#> 3 0.4_cwt 114.41044 6.887489 Estimable
#> 4 0.6_cwt 123.22901 6.912904 Estimable
#>
#> $avsed
#> overall
#> 3.748427
anova_table
Show an ANOVA table for multiple models, with p-values for each effect against each response.
rats[["newresp"]] <- rats[["weight"]] * runif(nrow(rats))
mod_rats2 <- asreml(
fixed = newresp ~ littersize + Sex + Dose,
random = ~idv(Dam),
residual = ~units,
data = rats,
trace = FALSE
)
mod_rats2 <- update(mod_rats2, aom = TRUE)
anova_table(mod_rats, mod_rats2)
#> Effect weight newresp
#> 1 littersize 0 0.804
#> 2 Sex 0 0.208
#> 3 Dose 0 0.008
pred_table
Get the mean for each level of a fixed effect, as well as standard error, and confidence intervals.
pred_table(mod_rats, classify = "Dose")
#> Error:
#> ! Classify must be type characterLeast significant difference
lsd_table
Show a least significant difference table:
lsd_table(mod_rats, classify = "Dose")
#> treatment group lsd means
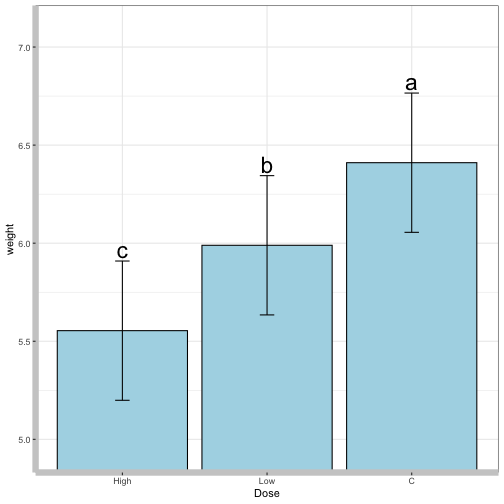
#> 1 C a 0.354997 6.410370
#> 2 Low b 0.354997 5.989315
#> 3 High c 0.354997 5.554064
lsd_graph
Show an LSD graph of the same data:
lsd_graph(mod_rats, classify = "Dose")